The Gaulton lab is a biomedical research group in the Department of Pediatrics and the UCSD Pediatric Diabetes Research Center at the University of California, San Diego School of Medicine in La Jolla, CA.
Our research interests include mapping gene regulatory programs involved in type 1 diabetes (T1D) and other complex diseases, modulating the activity of gene regulatory elements as therapies to prevent or treat T1D, and identifying novel biomarkers and improving risk prediction of T1D.
Research Areas
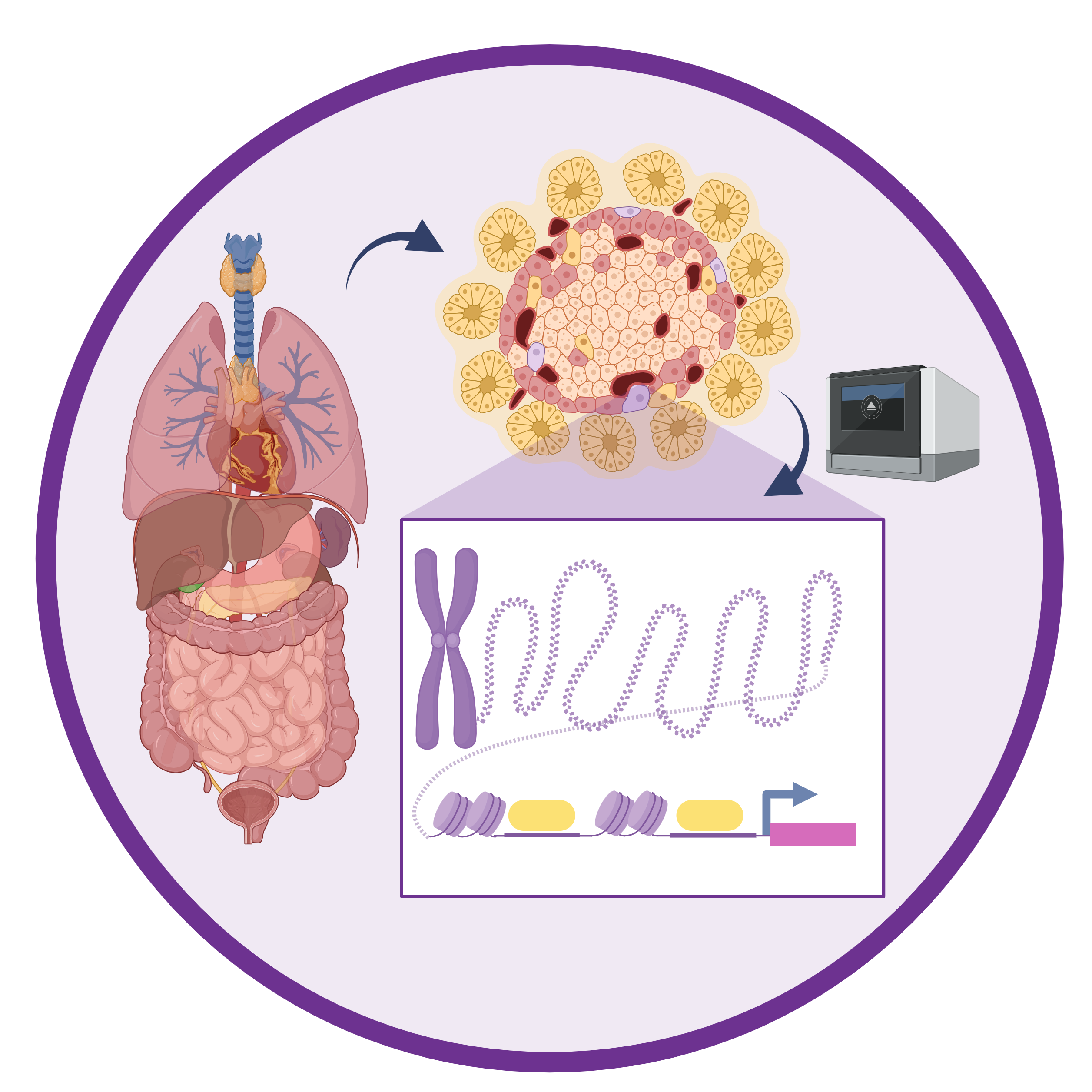
Single cell epigenomics of human tissues
Because the vast majority of disease-causing variants lie in non-coding regions of the human genome, understanding gene regulation is critical for understanding disease risk. Our lab is particularly interested in regions of the genome which influence the expression of nearby genes, aka cis-regulatory elements (CREs), and the regulatory networks they create. We have many completed and ongoing projects which use single cell methods to characterize the epigenome of human tissues including the pancreas, lungs, heart, liver, peripheral blood mononuclear cells (PBMCs), and more.
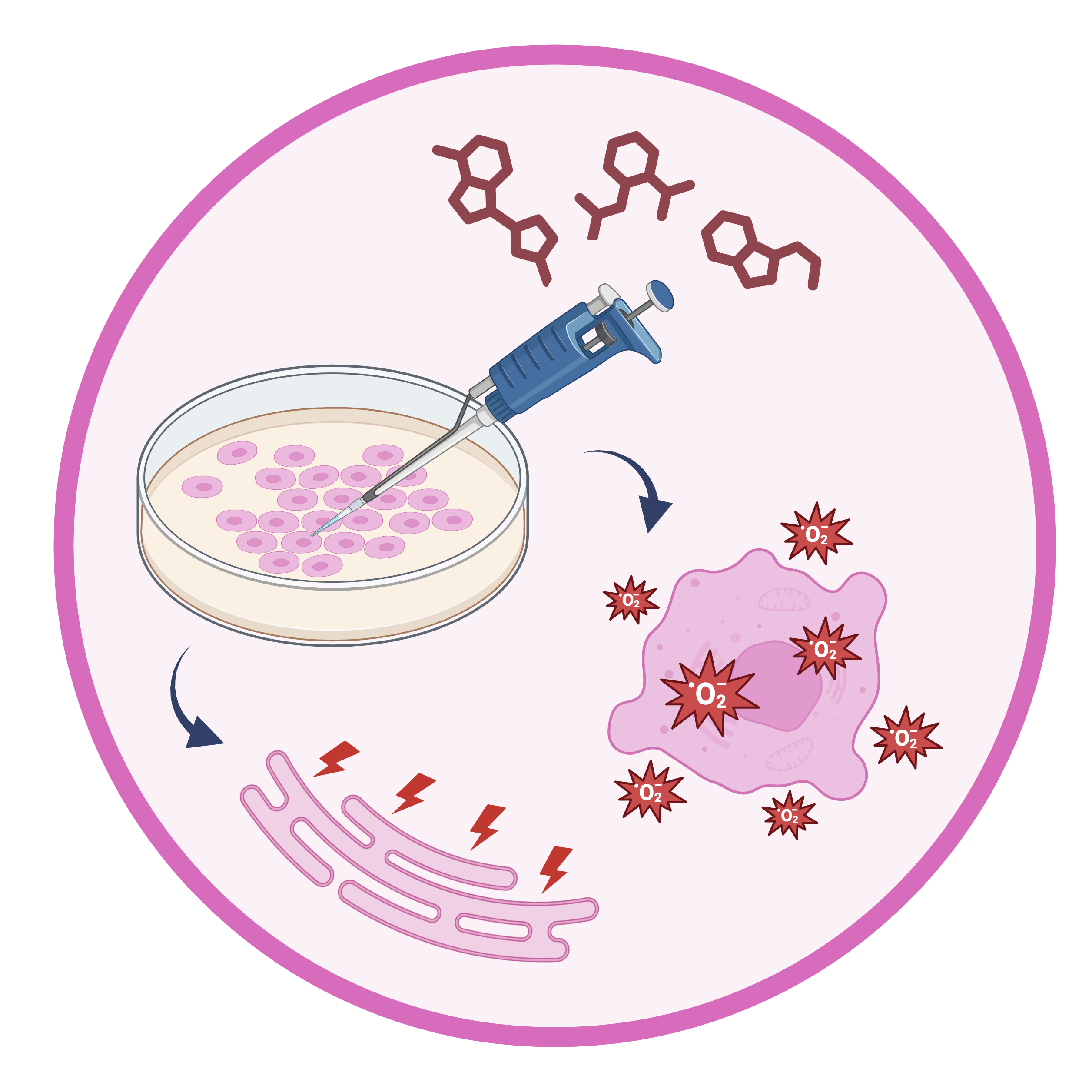
In vitro models of disease
Environmental stressors are predicted to play a role in the pathogenesis of diabetes, but their effects on the islet transcriptome and epigenome are largely unknown. Our lab uses a variety of small molecules and proteins to perform in vitro modeling of environmental stressors that pancreatic beta cells might be subject to, such as oxidative or ER stress. Downstream genetic analyses (scRNA-seq, snATAC-seq, etc.) help us to understand the effects of these stressors and how they relate to diabetes risk in humans.
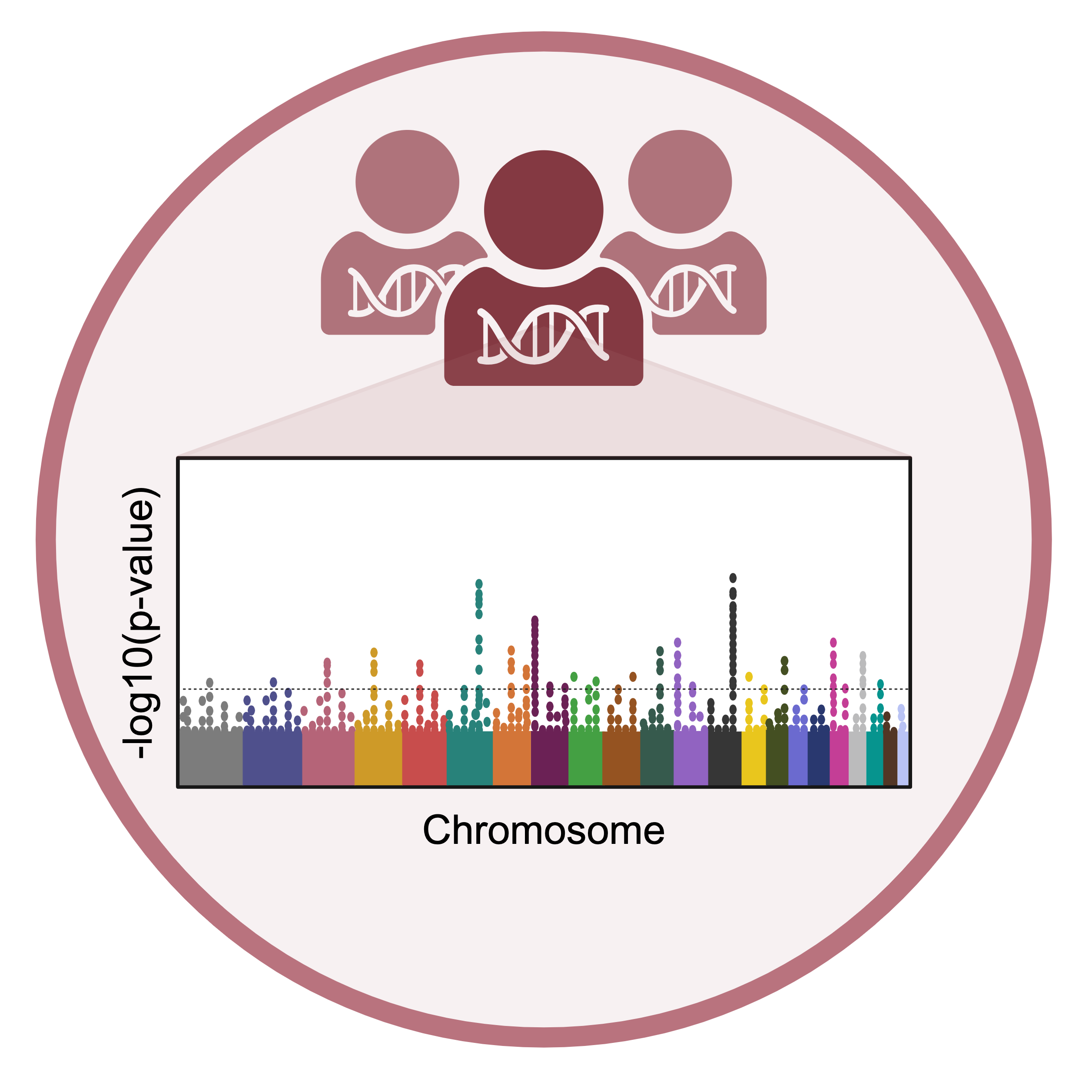
Population genetics
Diabetes is the most common metabolic disease worldwide. Our lab takes a population genetics approach to understanding underlying genetic risk in both type 1 and type 2 diabetes. We previously generated the largest genome-wide association study (GWAS) for type 1 diabetes to date, and our recent work in this area includes generating polygenic risk scores for disease along with quantitative trait loci mapping, particularly in human pancreatic islets.
Resources and Tools
Our lab takes part in several collaborative efforts to improve access and availability of scientific data and analytical pipelines. Our lab maintains key resources such as PanKBase, the Common Metabolic Diseases Genome Atlas (CMDGA), and the Knowledge Portal. Additionally, we aim to create resources and tools for the community to engage with our lab’s primary data.
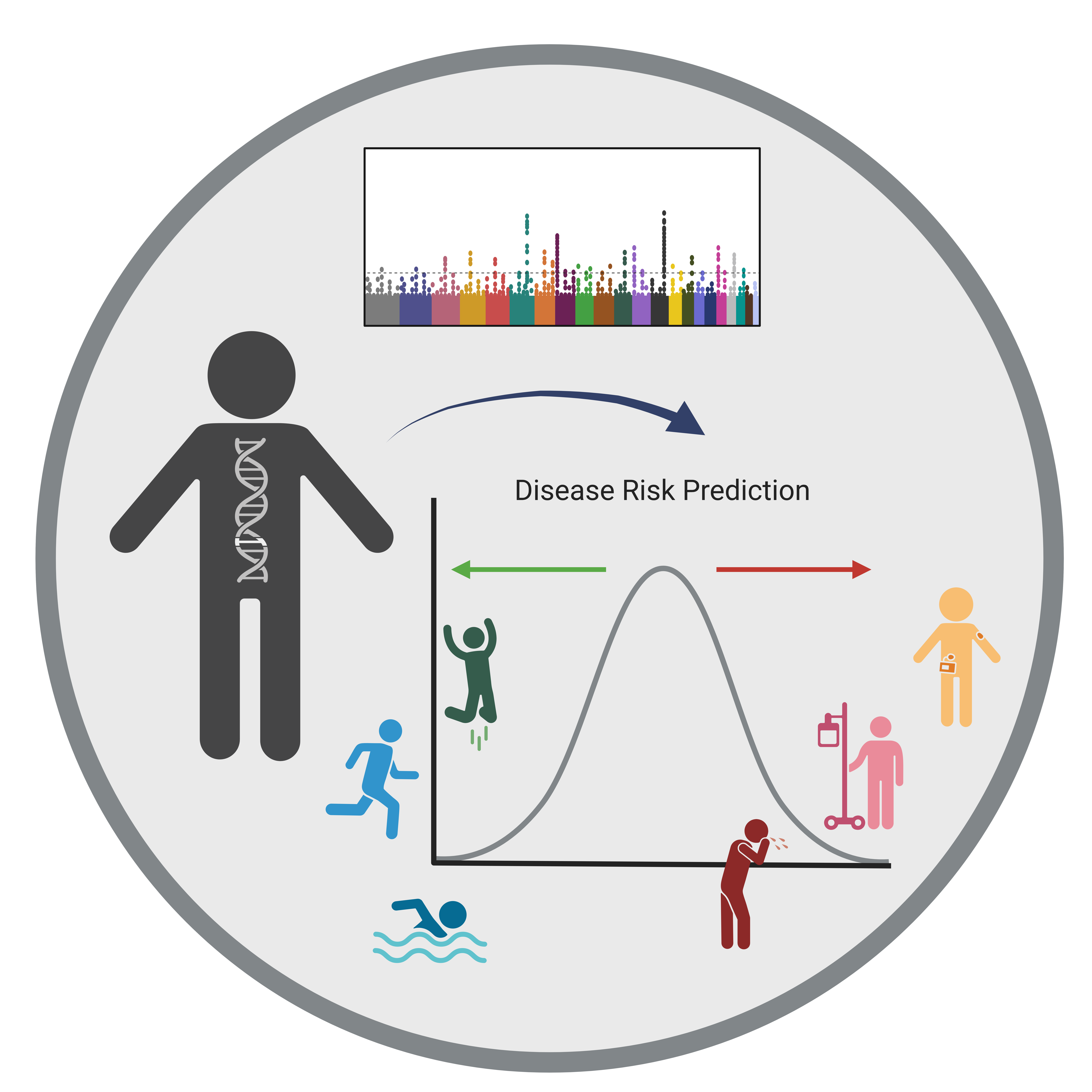
Disease risk prediction
Accurately predicting disease risk is crucial for preventing complications, guiding the selection of preventative therapies, and enhancing population-wide screening. Our lab aims to improve risk prediction in complex diseases, such as type 1 diabetes, by generating genetic risk scores (GRS) and identifying causal biomarkers that can be used to inform personalized medicine and improve early detection of disease. Our recent work includes developing a machine learning GRS and discovering novel metabolite biomarkers that significantly improve the accuracy of risk prediction.
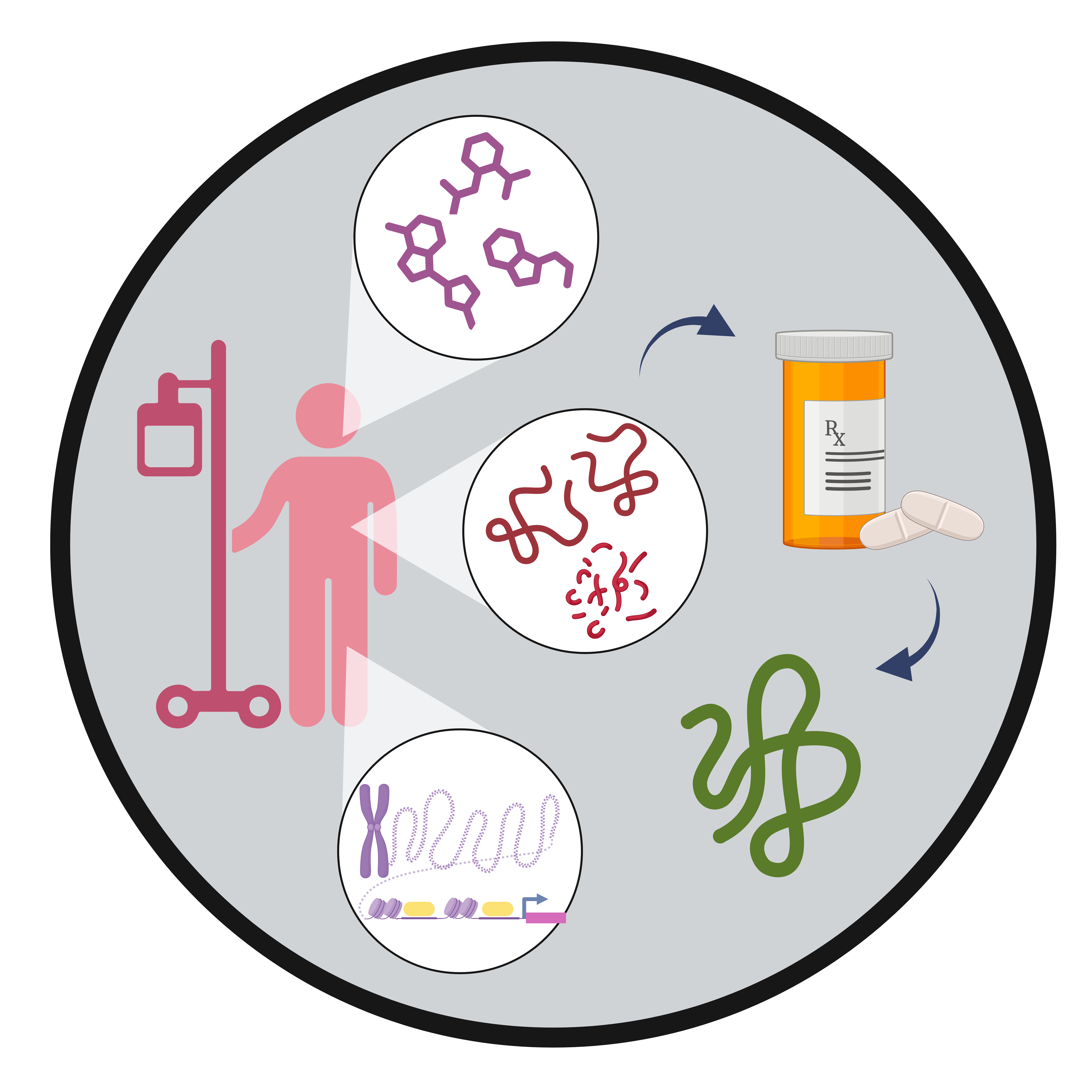
Identification of therapeutic targets
All of our lab’s work is towards the goal of understanding complex human diseases so we can ultimately prevent and treat them. We identify genes, cis-regulatory elements (CREs), proteins, and metabolites that change in individuals with diabetes and other diseases or are related to genetic and environmental risk of developing these diseases. All of these represent potential targets that could be modulated to prevent or treat disease.